Updating a phylogenetic tree
Interface
MolecularEvolution.AbstractUpdate Type
Summary
abstract type AbstractUpdate <: Function
A callable type that typically takes (tree::FelNode, models; partition_list=1:length(tree.message)), updates tree and models, and returns the updated tree and models.
Example
Define a new subtype, where foo and bar are arbitrary updating functions
struct MyUpdate <: AbstractUpdate end
function (update::MyUpdate)(tree::FelNode, models; partition_list=1:length(tree.message))
tree, models = foo(tree, models, partition_list=partition_list)
tree, models = BayesUpdate(nni=0)(tree, models, partition_list=partition_list)
tree, models = bar(tree, models, partition_list=partition_list)
return tree, models
endSee also: StandardUpdate
MolecularEvolution.StandardUpdate Type
Summary
struct StandardUpdate <: AbstractUpdate
A standard update can be a family of calls to nni_update!, branchlength_update!, root_update!, and model updates.
Constructor
StandardUpdate(
nni::Int,
branchlength::Int,
root::Int,
models::Int,
refresh::Bool,
nni_selection::Function,
branchlength_modifier::UnivariateModifier,
root_update::RootUpdate,
models_update::ModelsUpdate
)Arguments
nni::Int: the number of times to update the tree bynni_update!branchlength::Int: the number of times to update the tree bybranchlength_update!root::Int: the number of times to update the tree byroot_update!models::Int: the number of times to update the modelrefresh::Bool: whether to refresh the messages in tree between update operations to ensure message consistencynni_selection::Function: the function that selects between nni configurationsbranchlength_modifier::UnivariateModifier: the modifier to update a branchlength bybranchlength_update!root_update::RootUpdate: updates the root byroot_update!models_update::ModelsUpdate: updates the model parameters
See also: BayesUpdate, MaxLikUpdate
Example
using MolecularEvolution, Plots, DistributionsSimulate a tree
tree = sim_tree(n = 50)
initial_message = GaussianPartition()
models = BrownianMotion(0.0, 1.0)
internal_message_init!(tree, initial_message)
sample_down!(tree, models)
log_likelihood!(tree, models)-53.8373989462017Add some noise to the branch lengths
for n in getnodelist(tree)
n.branchlength += 100 * rand()
end
log_likelihood!(tree, models)-165.0275491877687Optimize under the brownian motion model
update = MaxLikUpdate(branchlength = 1, nni = 0, root = 1)
tree, models = update(tree, models)
@show log_likelihood!(tree, models)-23.02260454032541Set up a Bayesian model sampler
Let's assume the target of inference is not the tree itself, but rather the models. Assume further that you want to, for a fixed mean drift, sample the variance of the brownian motion model, with the metropolis algorithm. We begin with a struct that defines the model and how it's updated
tree = sim_tree(n = 200)
internal_message_init!(tree, GaussianPartition())
#Simulate brownian motion over the tree
models = BrownianMotion(0.0, 2.0)
sample_down!(tree, models)
mutable struct MyModelSampler{
T1<:ContinuousUnivariateDistribution,
T2<:ContinuousUnivariateDistribution,
} <: ModelsUpdate
acc_ratio::Tuple{Float64, Int64, Int64}
log_var_drift_proposal::T1
log_var_drift_prior::T2
mean_drift::Float64
function MyModelSampler(
log_var_drift_proposal::T1,
log_var_drift_prior::T2,
mean_drift::Float64,
) where {T1<:ContinuousUnivariateDistribution, T2<:ContinuousUnivariateDistribution}
new{T1, T2}((0.0, 0, 0), log_var_drift_proposal, log_var_drift_prior, mean_drift)
end
endThen we let this struct implement our metropolis_step interface
MolecularEvolution.tr(::MyModelSampler, x::BrownianMotion) = log(x.var_drift)
MolecularEvolution.invtr(modifier::MyModelSampler, x::Float64) =
BrownianMotion(modifier.mean_drift, exp(x))
MolecularEvolution.proposal(modifier::MyModelSampler, curr_value::Float64) =
curr_value + rand(modifier.log_var_drift_proposal)
MolecularEvolution.log_prior(modifier::MyModelSampler, x::Float64) =
logpdf(modifier.log_var_drift_prior, x)Now we define what a model update is
function (update::MyModelSampler)(
tree::FelNode,
models::BranchModel;
partition_list = 1:length(tree.message),
)
models = metropolis_step(update, models) do x::BrownianMotion
log_likelihood!(tree, x)
end
log_likelihood!(tree, models, partition_list = partition_list) #refresh messages
return models
endNow we define how the model is collapsed to its parameter
function MolecularEvolution.collapse_models(::MyModelSampler, models::BranchModel)
return models.var_drift
endNow we define a Bayesian sampler
update = BayesUpdate(
nni = 0,
branchlength = 0,
models = 1,
refresh = true,
models_sampler = MyModelSampler(Normal(0.0, 1.0), Normal(-1.0, 0.2), 0.0),
)
trees, LLs, models_samples = metropolis_sample(
update,
tree,
BrownianMotion(0.0, 7.67),
1000,
burn_in = 1000,
collect_LLs = true,
collect_models = true,
)
ll(x) = log_likelihood!(tree, BrownianMotion(0.0, x))
prior(x) = logpdf(update.models_update.log_var_drift_prior, log(x)) - log(x)
x_range = 0.1:0.1:5
p1 = histogram(
models_samples,
normalize = :pdf,
alpha = 0.5,
label = "Posterior samples",
xlims = (minimum(x_range), maximum(x_range)),
xlabel = "variance per unit time",
ylabel = "probability density",
)
p2 = plot(x_range, ll, label = "Tree likelihood")
p3 = plot(x_range, prior, label = "Prior")
plot(p1, p2, p3, layout = (1, 3), size = (1100, 400))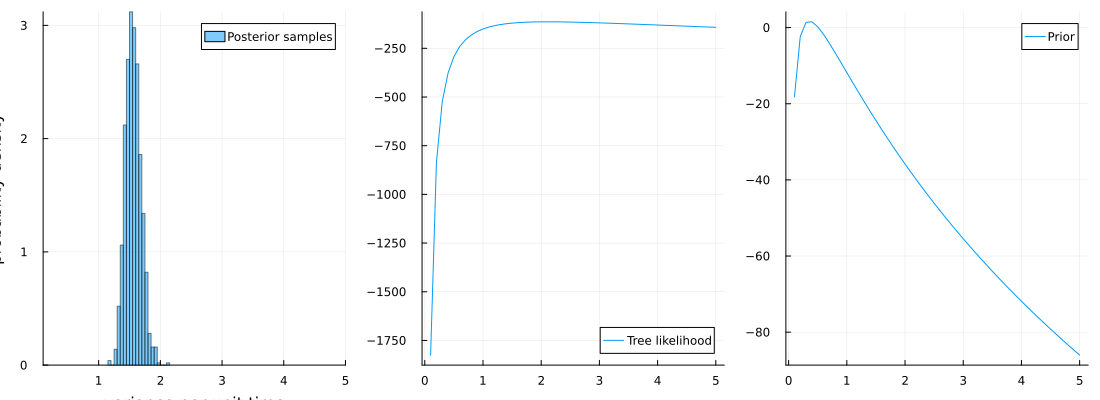
plot(LLs)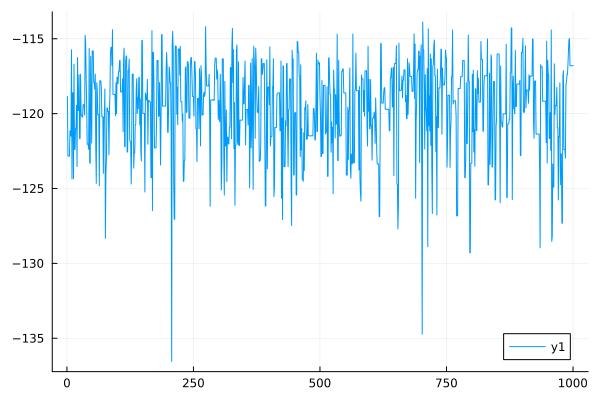
plot(models_samples)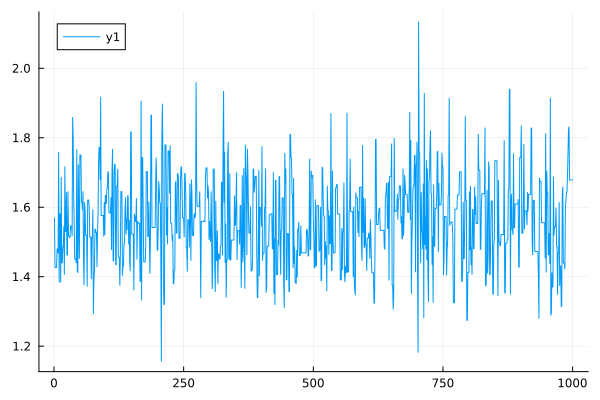
This page was generated using Literate.jl.